NEBcutter: a program to cleave DNA with restriction enzymes
NEBcutter V2.0 is a program that can take a input DNA sequence and find large (maximum sequence length of 300kb), non-overlapping open reading frames and the sites for all restriction enzymes that cut the sequence just once. It also shows the enzymes that could be used in a complete digest to excise each open reading frame that it finds. NEBcutter incorporates everything that is known about the methylation sensitivity of any of the enzymes displayed when they overlap Dam or Dcm sites as well as CpG, EcoK and EcoB sites. Made available on the Web by New England Biolabs, Inc (http://tools.neb.com/NEBcutter) . It produces a variety of outputs including restriction enzyme maps, theoretical digests and links into the restriction enzyme database, REBASE (http://www.neb.com/rebase).
Using NEBcutter
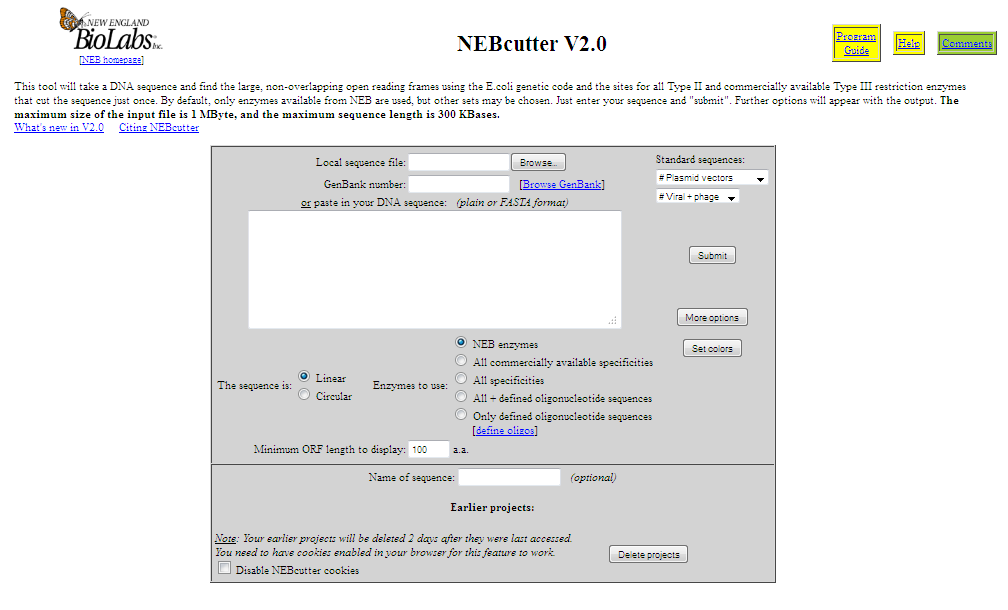
NEBcutter accepts an input sequence, which can be pasted in, picked up from a local file or retrieved from NCBI as a GenBank file via its accession number. Various options are available to select the size of ORFs to be displayed and the set of restriction enzymes to be used. The program calculates the positions of all restriction enzyme sites noting those that might potentially be blocked by overlapping methylation and finds the ORFs in the sequence. It then displays a schematic diagram of the sequence, the long ORFs, based on the rules described in the Methods and all restriction enzymes that cut it just once. The initial display also shows the enzymes that can be used in a complete digest to excise each ORF that is displayed. Figure 1 shows a typical digest, in this case a linear view of the plasmid pBR322. If the original DNA sequence is circular, then both linear and circular displays are offered. From the initial display there are many options to go further, including custom digestion with enzymes of your choice and various displays of the digest. It is also possible, from linear displays, to zoom in on a selected region of the sequence for a more detailed view. One option allows a particular region to be selected and those enzymes that cut immediately adjacent to the region to be displayed. This is useful to find enzymes able to excise the desired region. If zooming leads to a region of <60 nucleotides then the actual sequence is displayed (Fig. 2), together with any translation that is appropriate. On this display all enzymes that cut the sequence are shown, all bases that form parts of a restriction enzyme recognition sequence are highlighted and moving the mouse over an enzyme name will produce a box with the recognition sequence noted and the sequence itself becomes underlined in the display.
This tool will take a DNA sequence and find the large, non-overlapping open reading frames using the E.coli genetic code and the sites for all Type II and commercially available Type III restriction enzymes that cut the sequence just once. By default, only enzymes available from NEB are used, but other sets may be chosen. Just enter your sequence and "submit". Further options will appear with the output. The maximum size of the input file is 1 MByte, and the maximum sequence length is 300 KBases. [19]
On any main display, moving the mouse over a restriction enzyme name will show its recognition sequence and the precise base number at which it cleaves. If more than one site is present for the enzyme then all other sites are underlined. Clicking on the enzyme name will produce a page with a summary of information about the enzyme including its methylation sensitivity, the kinds of ends produced, isoschizomers that are available and a list of other enzymes that can produce compatible ends. For NEB enzymes, information about digestion conditions and a link to the NEB web site is provided.
On every page hot links are provided that lead back to the main page, provide help in interpreting the display or allow users to send comments back to the authors. The interface has been designed to be as intuitive as possible and to provide easy access to the main functions useful to researchers attempting to cleave their DNA. The current version of NEBcutter has processed more than 400 000 sequences from users.Reference:
- http://nar.oxfordjournals.org/content/31/13/3688.full
- http://www.funfaculty.org/drupal/node/2491