pUC19 an example of plasmid vector
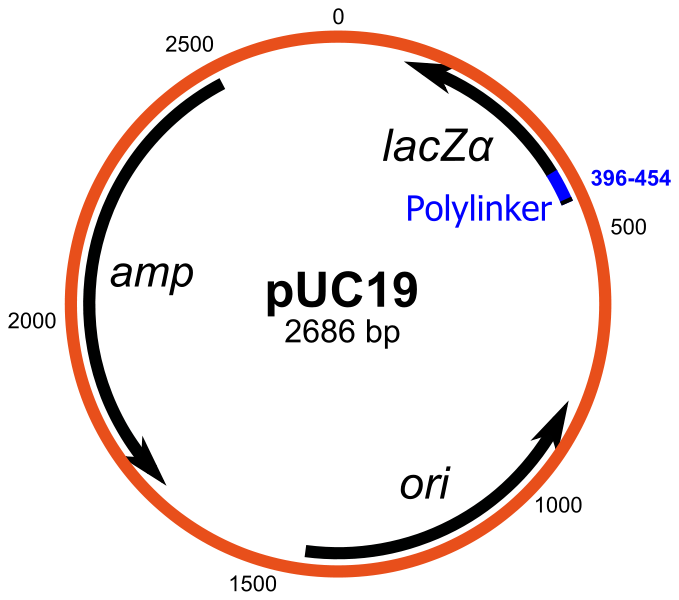
Image courtesy: http://upload.wikimedia.org/wikipedia/commons/thumb/8/8d/PUC19.svg/679px-PUC19.svg.png
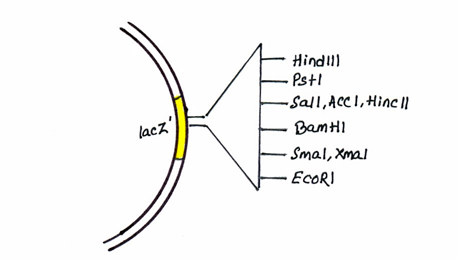
Image courtesy: http://www.discoverbiotech.com/image/image_gallery?uuid=2ae8f77e-77a6-4d5c-b755-50088228ca06&groupId=11406&t=1332224388076
The valuable features of pBR322 have been enhanced by the construction of a series of plasmids termed pUC (produced at the University of California). [5]
There is an antibiotic resistance gene for tetracycline and origin of replication for E. coli.[5] There is presence of multiple restriction sites termed the multiple cloning site (MCS).
It additionally codes for a portion of a polypeptide called β-galactosidase.
When the pUC plasmid has been used to transform the host cell E. coli the gene may be switched on by adding the inducer IPTG (isopropyl-β-D-thiogalactopyranoside).[5] Its presence causes the enzyme β-galactosidase to be produced. The functional enzyme is able to hydrolyse a colourless substance called X-gal (5-bromo-4-chloro-3-indolylb-galactopyranoside) into a blue insoluble material (5,50-dibromo-4,40–dichloro indigo). [5] However if the gene is disrupted by the insertion of a foreign fragment of DNA, a non-functional enzyme results which is unable to carry out hydrolysis of X-gal. [5] This makes the recombinant pUC plasmid to be easily detected as it is white or colourless in the presence of X-gal. an intact non-recombinant pUC plasmid will be blue since its gene is fully functional and not disrupted. This elegant system, termed blue/white selection, allows the initial identification of recombinants to be undertaken very quickly and has been included in a number of subsequent vector systems. [5]
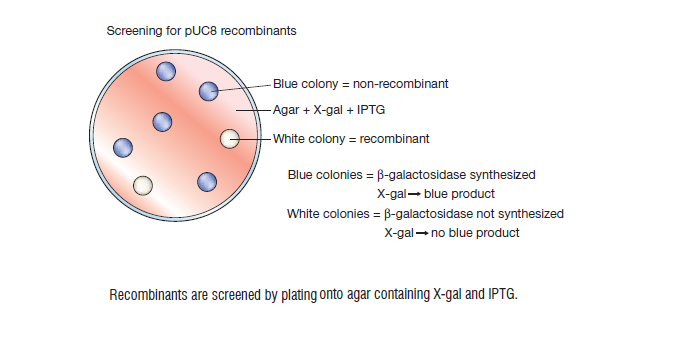
Image courtesy: Fig 5.10, Gene Cloning & DNA Analysis- An Introduction, T.A.Brown. fifth edition, blackwell publishing, 2010
PGEM3Z plasmid
pGEM3Z is having similarity with a pUC vector because of the presence of the ampR genes and lacZ' genes containing a cluster of restriction sites. The difference is that pGEM3Z has two additional, short pieces of DNA, each of which acts as the recognition site for attachment of an RNA polymerase enzyme. These two promoter sequences lie on either side of the cluster of restriction site used for introduction of new DNA into the pGEM3Z molecule. [24]
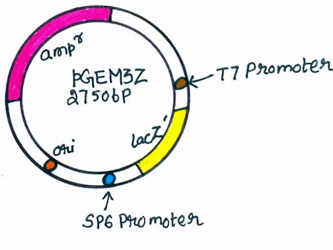
Image courtesy: http://www.discoverbiotech.com/image/image_gallery?uuid=0b023d89-83fa-4e37-a197-cd90aeb514d5&groupId=11406&t=1332224388079
Advantages:
- High copy number
- Identification can be achieved by single step by plating on agar medium contaning ampicillin and X gal.
- Time consumption is less.
- pUC vectors carry different combinations of restriction sites and show greater flexibility in the types of DNA fragment that can be cloned. Clustering of the restriction sites allows a DNA fragment with two different sticky ends to be cloned without involving linker attachment. DNA cloned into a member of the pUC series can be transferred directly to its M13mp counterpart, because of the same restriction site clusters and it can be analysed by DNA sequencing or in vitro mutagenesis.[24]
Reference:
- Principles and Techniques of Biochemistry and Molecular Biology , Wilson Keith and Walker John, seventh edition, cambridge university press, 2010
- Gene Cloning & DNA Analysis- An Introduction, T.A.Brown. fifth edition, blackwell publishing, 2010
- http://www.discoverbiotech.com/wiki/-/wiki/Main/Cloning+Vectors